
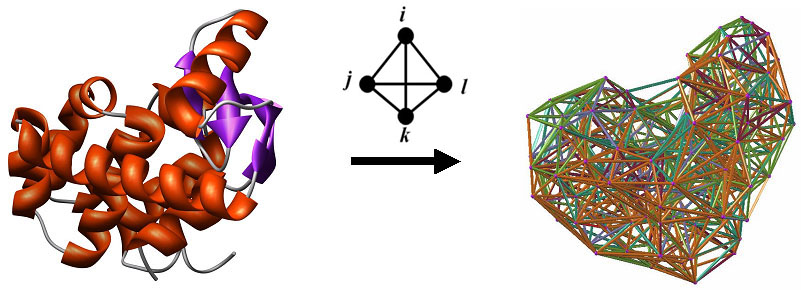
These structural change data are used as attributes to generate a vector representation for each nsSNP, in combination with additional features reflecting sequence and structure of the corresponding protein. In this study, we characterize a representative sampling of 1790 documented neutral and disease-related human nsSNPs mapped to 243 diverse human protein structures, by quantifying environmental perturbations in the associated proteins with the use of a computational mutagenesis methodology that relies on a four-body, knowledge-based, statistical contact potential. The nature of protein structure-function relationships suggests that nsSNP effects, either benign or leading to aberrant protein function possibly associated with disease, are dependent on relative structural changes introduced upon mutation. In particular, substantial interest exists in determining whether a non-synonymous SNP (nsSNP), leading to a single residue replacement in the translated protein product, is neutral or disease-related. N2 - Certain genetic variations in the human population are associated with heritable diseases, and single nucleotide polymorphisms (SNPs) represent the most common form of such differences in DNA sequence. T1 - Knowledge-based computational mutagenesis for predicting the disease potential of human non-synonymous single nucleotide polymorphisms. A dedicated server for obtaining predictions, as well as supporting datasets and documentation, is available at. Our classifier performs at least as well as other methods that use significantly larger datasets of nsSNPs for model training, and the novelty of our attributes differentiates the model as an orthogonal approach that can be utilized in conjunction with other techniques. A trained model based on the random forest supervised classification algorithm achieves 76% cross-validation accuracy.


Certain genetic variations in the human population are associated with heritable diseases, and single nucleotide polymorphisms (SNPs) represent the most common form of such differences in DNA sequence.


 0 kommentar(er)
0 kommentar(er)
